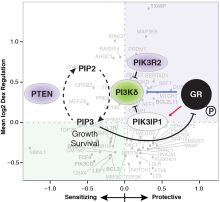
The childhood blood cancer B-cell precursor acute lymphoblastic leukemia (B-ALL) responds to treatment with dexamethasone in ~90% of patients, but there are few therapeutic options in children resistant to dexamethasone. The mechanism by which dexamethasone kills B-ALL cancer cells, and the determinants of response to therapy are unclear. In a collaboration lead by Dr. Miles Pufall (University of Iowa), we used our functional genomics platform to uncover genes controlling the response of the childhood cancer to the anti-cancer drug dexamethasone. Integrating the results from this genetic screen with transcriptomic data revealed that dexamethasone was effective against B-ALL by suppressing B cell development genes. It also uncovered a regulatory feedback loop involving Phosphoinositide 3-kinase delta (PI3Kδ). A combination therapy of dexamethasone with the PI3Kδ inhibitor idelalisib killed even highly resistant B-ALL cells and showed synergistic anti-cancer effects in a mouse model of B-ALL.
The results from our study are in press at Blood:
Kruth K, Fang M, Shelton DN, Abu-Halawa O, Mahling R, Yang H, Weissman JS, Loh ML, Müschen M, Tasian SK, Bassik MC, Kampmann M, Pufall MA (2017) Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. pii: blood-2017-02-766204. doi: 10.1182/blood-2017-02-766204. [Epub ahead of print]